- < Back To Home
- /
- How To & References
How To & References
Apply for AMP PD Access
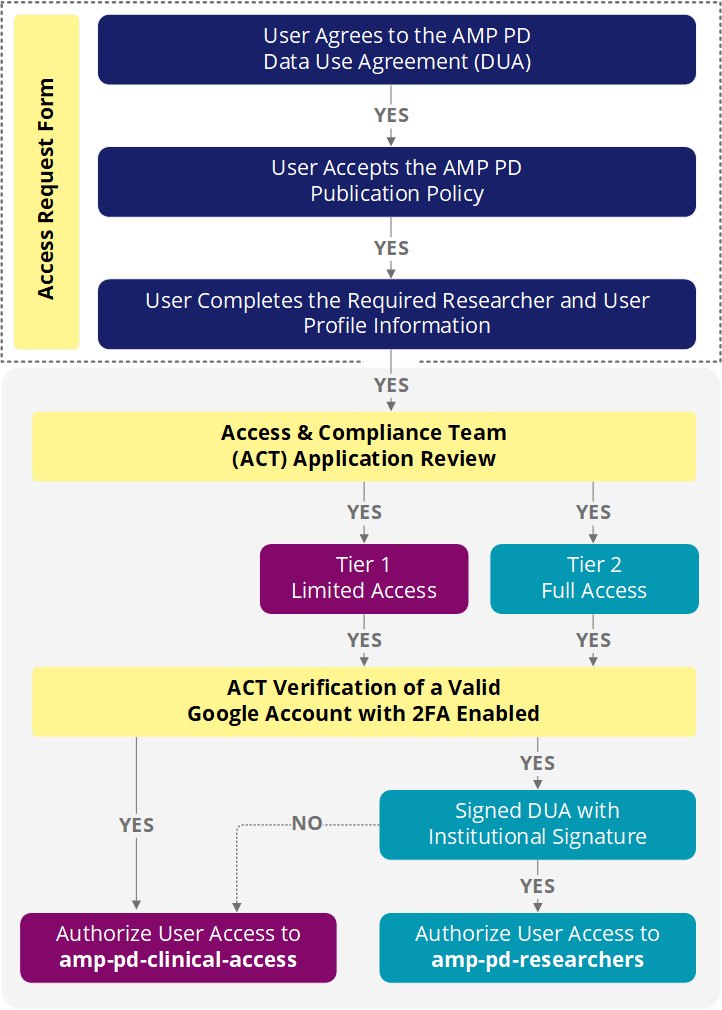
1. AMP PD Access Request Form
2. Setting up a Google Account
Access to AMP PD data and Terra requires a Google account. This Google Account can be:
- An institutional account, if your institution uses Google gSuite
- A Google account associated with your institutional email address
- A Gmail account
If your institutional email account is a Google account, please use it. If not, linking your institutional email address will allow you sign in, recover your password, and receive notifications via your primary/work email. To link your primary/work email address, reference the following steps:
Go to the Google sign-up page
- Under "Username", click "Use my current email address instead"
- Enter the email you want to use for you AMP PD account
- You will receive an email from Google Accounts. Verify your email address by inputting the security code included in the email
- Complete the rest of your profile information
Click the following link to access the full set of instructions for linking your primary/work email address to a Google account
3. Setting up 2-Step Verification
In accordance with AMP PD governance and Terra policy, users must provide a valid Google account and have 2-Step Verification activated. If you do not currently have a Google account or 2-Step Verification setup, you may still submit your AMP PD access request to the ACT for review and further coordination. To setup 2-Step Verification, reference the following steps:
- Go to your Google Account
- On the left navigation panel, click Security
- On the Signing in to Google panel, click 2-Step Verification
- Click Get Started
- Follow the steps on the screen
Click the following link to access the full set of instructions for setting up 2-Step Verification
4. Requirements for accessing genomics data
Users who want to access AMP PD genomics data ("Full Access" permissions) must provide a signature from an authorized Institutional Signing Official via the AMP PD Data Use Agreement (DUA). An Institutional Signing Official is generally a senior official at an institution who is credentialed through NIH eRA Commons system and is authorized to enter the institution into a legally binding contract and sign on behalf of an investigator who has submitted data or a data access request to NIH.
You can download, sign, and upload the signed version of the DUA when submitting your application. Alternatively, you can coordinate with a member of the AMP PD Access & Compliance Team (ACT) to provide this required information.
Please note that failure to provide an institutional signature will restrict access to all AMP PD research data. By default, users will be assigned permissions to the "Limited Access" (clinical and summary data) tier until an institutional signature and signed DUA is provided.
5. Full AMP PD Application Submission and Review Process
Citations and References
Tracking of Data Use and Publications for Registered AMP PD Knowledge Platform users
If AMP PD data are used in publications, authors will be required to:
- Add AMP PD acknowledgement (see Data Use Agreement)
- Add AMP PD cohort(s) acknowledgement(s) if data used is derived from the cohorts represented in AMP PD (see Data Use Agreement) Include recommended language describing AMP PD methods and data gathering
- Cite AMP PD sponsors (see Data Use Agreement)
- Provide manuscript to AMP PD publication coordination team for review 2 weeks prior to planned submission (see Data Use Agreement)
- Apprise AMP PD publication coordination team of acceptance or rejection at manuscript@amp-pd.org
- Provide manuscript citation to AMP PD publication coordination team upon acceptance at manuscript@amp-pd.org
- Provide URL to published work, if possible.
Contribute Data
Contributing study data benefits your study and AMP PD. When you contribute your clinical data, it will be harmonized with existing AMP PD data to facilitate search and discovery. This also provides your study greater exposure with links back to your study as the source of truth for detailed clinical data. Current AMP PD cohorts include clinical data and at least one set of DNA, RNA, and Proteomic assay data. When you contribute this data, your study data will be integrated into the AMP PD sample catalog using a standardized nomenclature and will enjoy the benefits of a wealth of AMP PD developed analyses and versioned pipeline workflows.
The following criteria describe considerations for determining whether data from a Study is suitable for inclusion in AMP PD. A sample of considerations for including a genetic study is also described.
New cohort criteria
- Data Agreements to share data with AMP PD in place
- Relevance to PD research
- Size (cases and controls)
- Patient consent should include approval for broad data-sharing of de-identified clinical and molecular data
- Study must be able to fall under AMP PD DUC
- Longitudinal study (frequency and standardization of visits)
- Clinical data (demographics, family history, medicine, UPDRS/MDS-UPDRS, MoCA, MMSE, UPSIT, ESS, Epworth)
- Biosample availability, sample collection matches AMP PD cohorts, QC available
- Pathological sample data must be broadly shareable
Genetic Studies
- Relevance to PD research
- Size (cases and controls)
- Patient consent should include approval for broad data-sharing of de-identified clinical and molecular data
- Clinical data (demographics, family history, medicine, UPDRS/MDS-UPDRS, MoCA, MMSE, UPSIT, ESS, Epworth)
- Pathological data (definition of pathology, immunoassays performed, quantitative or descriptive, standard criteria)
- Cost-benefit analysis regarding storage and analytical costs
- If a genetic study meets the criteria, then an additional question is “who would run GATK pipeline, CRAM validation, and joint calling?”
SC members expressed support for these criteria and emphasized the importance of ensuring seamless access by researchers through a single data use agreement.
In applying these criteria, the SC agreed that adding the LBD cohort is a good starting point because (1) AMP PD already has the pipeline in the cloud and knows how to run it, (2) LBD is on the PD continuum, and (3) the cases have been pathologically confirmed.
Example of Genetic Study - Lewy Body Dementia
- Lewy Body Dementia (LBD - 2nd most common form of dementia -1.4 M in US)
- Genetic defects are common in LBD, but poorly understood
- Pathological samples - neocortical or limbic DLB, age, sex, race, disease, subtype
- Clinical samples - age, sex, race, disease, subtype
- 3000 cases: 75% pathological, 25% clinical (from PDBP LBD cohort)
- 2,000 controls from Baltimore longitudinal study on aging, NCRAD - balance proportion of male/female with cases
- Illumina X10 platform 150 by, paired-end, 30X coverage, single library, single lane clustering
- Data freeze Dec 2018, variant calls Feb 2019 (Build 38)
- Plan to also do joint call with TopMED
- Early analysis (contamination, sex chromosome ratio, heterozygous/homozygous)
To discuss the contribution of your study data to AMP PD, please email admin@amp-pd.org, to arrange a discussion on this exciting opportunity with key AMP PD stakeholders.
Data Access Types
AMP PD study data is controlled access data. Only individuals who have been approved for access by the program should have access.
AMP PD data access is controlled using a two (2) tier model:
- Tier 1: Limited Access - Access to participant-level clinical data, along with limited information about genomic and transcriptomic samples. This includes lists of participant samples and can include aggregated information about genomic mutations.
- Tier 2: Full Access - All AMP PD research data, including participant-level clinical, genomic, and transcriptomic data and metadata
Limited Access |
Full Access |
|
| Row-Level Clinical Data | ||
| Participant Table | ||
| Case/Control Table | ||
| WGS Inventory Metadata | ||
| RNA Inventory Metadata | ||
| Getting Started Work-space | ||
| WGS Samples, Joint Genotyping & Plink Files | ||
| RNA Samples & Plink Files | ||
| WGS & RNA Specific Work-spaces | ||
| WGS & RNA BigQuery Tables | ||
| Targeted Proteomics Preview |
For those requesting access to all AMP PD data (Full Access), please provide an institutional signature verifying that their host institution adopts responsibility for appropriate use of the data and for the security of any downloaded data.
Data Download Inquiry & Justification
In order to create the best platform possible for sharing and collaborating in research, including best practices for data security, AMP PD encourages cloud analysis and seeks to understand use cases for data downloads. We would like to hear from you as to why you may want to download data. Please contact the ACT prior to downloading data at: ACT@amp-pd.org.
STRIDES & Setting Up Cloud Credits
The NIH Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) Initiative allows NIH to explore the use of cloud environments to streamline NIH data use by partnering with commercial providers such as Google Cloud and Amazon Web Services (AWS). NIH’s STRIDES Initiative provides cost-effective access to industry-leading partners to help advance biomedical research.
Specific to AMP PD users, the STRIDES initiative provides the following benefits:
- Discounts on STRIDES Initiative partner services—Favorable pricing on computing, storage, and related cloud services for NIH Institutes, Centers, and Offices (ICOs) and NIH-funded institutions. For AMP PD users, access to STRIDES discounts can be achieved by one of the following methods:
- Setting up an AMP PD billing account. This is appropriate for AMP PD data generation, processing, and consortia approved analysis
- Creating an AMP PD billing sub-account
- Establishing an agreement between your institute(s) and STRIDES. This is applicable to NIH funded researchers and institutes.
- Professional services—Access to professional service consultations and technical support from the STRIDES Initiative partners.
- Training—Access to training for researchers, data owners, and others to help ensure optimal use of available tools and technologies.
- Potential collaborative engagements—Opportunities to explore methods and approaches that may advance NIH’s biomedical research objectives (with scope and milestones of engagements agreed upon separately).
At this time, the STRIDES Initiative supports programs/projects who want to prepare, migrate, upload, and compute on data in the cloud. In the future, the ability to access data across NIH and NIH-funded institutions from various research domain repositories will become available. For contact information or for additional details on STRIDES please visit the NIH STRIDES Initiative website.
Sign up for Google Cloud Billing
Because Terra runs on the Google Cloud Platform, all Terra costs, including storage, compute, and data egress, are ultimately billed via Google Billing Accounts.
You will need to provide billing information, but can start with a free trial that includes $300 in compute and storage credits. Click here for additional details regarding free credits.
In order to be compatible with multiple cloud environments, institutional payment systems, and security requirements, the Terra interface does not directly display the Google Cloud Console Billing interface. Instead, Terra will connect a Google Billing Account to an associated Terra Billing Project. When using Terra, it is the Terra Billing Projects that you will see in the interface, and to which Terra will charge your usage costs. In order to create or clone a new workspace in Terra, you must have access to at least one Terra Billing Project.
For additional information, Click here.