- < Back To Home
- /
- Cloud Cost Guidelines for AMP PD
Cloud Cost Guidelines for AMP PD
Cloud Cost Overview
Data for AMP PD is made available on the Google Cloud Platform (GCP). This data is stored in Google Cloud projects paid for by the AMP PD partnership.
To use AMP PD data, you must have a Google Cloud billing account, and any charges related to using the data or Google Cloud services will be billed to you by Google. This document provides information and examples for working with AMP PD data so that you can make informed decisions around costs. For up-to-date billing information, see the documentation for GCP Pricing.
1. Platform Elements & Architecture
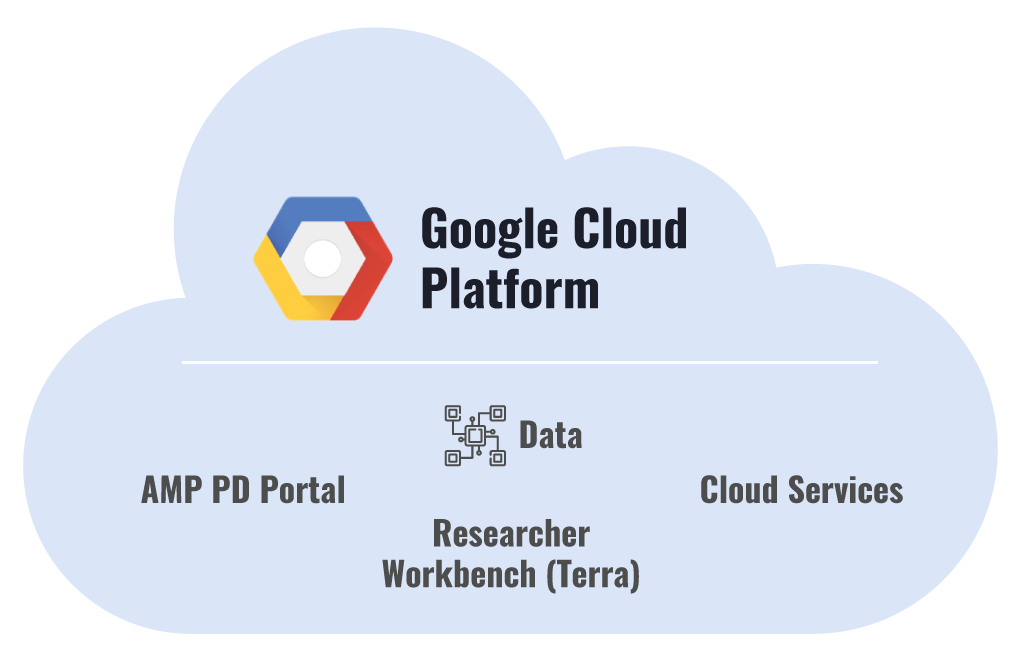
Key elements of the platform include:
- AMP PD Portal
- Cloud Services
- Researcher Workbench (Terra)
- AMP PD Data
2. AMP PD Portal
The AMP PD Portal is a web site that provides information about the AMP PD project, such as its mission, the partnership, and the available data. There is no cost for accessing the AMP PD Portal.
3. Cloud Services
AMP PD data is made available on the Google Cloud Platform (GCP) such that researchers have the freedom to use all GCP services. Use of GCP services will incur charges as documented at cloud.google.com.
4. Researcher Workbench (Terra)
The Researcher Workbench (also known as Terra), is a platform focused around biomedical research. Terra itself is free to use, however specific operations in Terra, including running Workflows, running Notebooks, and accessing and storing data, may incur Cloud charges.
5. AMP PD Data
Data for AMP PD is stored in Google Cloud projects paid for by the AMP PD partnership. You do not incur charges for storing AMP PD data unless you make a copy of it. You may incur charges when accessing AMP PD data.
AMP PD data in Google Cloud Storage is stored in Requester Pays Buckets in the region us-central1. To access this data, you must have a Google Cloud project (such as a Terra Billing Project) to bill to.2
Light listing or browsing of data will not generate charges. Accessing data from Compute Engine VMs (such as a Terra Notebook Runtime VM) running in us-central1 will not incur charges.
Downloading data or accessing data from Compute Engine VMs in regions other than us-central1 will incur charges as described in the documentation on egress charges.
6. Common Use Cases
Subsequent discussion of Cloud costs are intended to address cost considerations for these use cases.
- Running a workflow
- Converting a CRAM to a BAM
- Aligning a genomic sample to a reference and performing variant calling using the GATK
- Aligning a transcriptomic sample to a reference using STAR
- Running a notebook
- Performing Quality Control checks on genomic data
- Analyzing genomic variants
- Storing files in Cloud
- Notebooks
- Clinical Data
- Genomic Data
- Transcriptomics Data
7. Cloud Concepts
As relates to AMP PD, cloud costs fall into the following general categories:
- Storage
- Compute and disks
- Query processing
- Data egress
- Data retrieval
As relates to AMP PD, the key cloud services used are:
- Google Cloud Storage
- Google Compute Engine
- Google BigQuery
Google Cloud Storage
Google Cloud Storage (GCS) is an "object store" where "objects" are stored in "buckets". More commonly, one can think of it as a place to store files in a structure similar to "folders" or "directories". For more details, see How Sub-directories Work.
1. Google Cloud Storage Overview
AMP PD genomic data is stored in a bucket named amp-pd-genomics, and a CRAM file for a sample can be found in the path:
gs://amp-pd-genomics/samples/wgs/gatk/<sample-id>/<sample-id>.cram
Storage has a cost associated with it. In addition when you access data in GCS, you want to consider where the data will be accessed from as moving data out of GCS may incur charges.
The framework for storage costs is:
- How much are you storing?
- Where are you storing it and how frequently is it accessed?
2. How much are you storing?
A key cloud concept is that you only pay for what you use. Thus you don't need to pre-allocate storage in GCS (like buying an array of disks); you simply pay for what you store.
3. Where are you storing it and how frequently is it accessed?
Google Cloud Storage provides several different storage classes, each with different pricing. The options are primarily based around where you want to store the data and how frequently you will access the data.
Data that is stored in more locations (multiple "regions") is more expensive than data that is stored in fewer locations ("regional"). For more information, you can read about Google Cloud regions and bucket locations.
Data that will be accessed the most frequently should be stored in a more expensive storage tier. Data that will be accessed infrequently can be stored in less expensive "cold" storage.
4. Storage Classes - Multi-Regional and Regional Storage
Multi-Regional storage is the most expensive option at $0.026 per GB per month. Regional storage is less expensive at $0.020 per GB per month.
Multi-Regional storage is most appropriate for data that needs to be accessed quickly and frequently from many locations (for a web site or for gaming, for example).
This is not typically the case for genomic or transcriptomic research data. With these data types, overall access frequency is low and emphasis is on managing storage costs. AMP PD data published to researchers is stored in Regional GCS buckets in us-central1.
5. Storage Classes - Nearline and Coldline Storage
For data that will be accessed very infrequently, Google Cloud offers Nearline and Coldline storage. These storage classes offer significantly reduced costs for storage ($0.010 per GB for Nearline and $0.007 per GB for Coldline), but add a retrieval charge ($0.01 per GB and $0.05 per GB for Coldline).
These storage classes are most appropriate for archive data, for example, after processing FASTQs into BAMs or CRAMs.
6. GCS - Egress Costs
Egress charges apply when copying GCS data out of the region(s) that the data is stored in. For example:
- Downloading data to your workstation or laptop
- Copying data stored in one region to a Compute Engine VM in another region
- Copying data stored in one region to a GCS bucket in another region
- Copying data stored in a multi-region bucket to a regional GCS bucket
Network egress charges vary, but within the United States, the cost is typically $0.01 per GB.
If GCS data is accessed from within the same Cloud region in which the data is stored, no egress charges will be incurred. For example, accessing AMP PD data (stored in us-central1) using a Google Compute Engine VM running in us-central1-a will incur no egress charges.
7. GCS - Retrieval costs
Retrieval costs apply only to the "cold storage" classes, Nearline and Coldline. Note that retrieval applies to:
- Copying data from a cold storage bucket
- Moving data within a cold storage bucket (a move is a copy followed by a deletion)
Google Compute Engine
Google Compute Engine (GCE) provides virtual machines (VMs) and block storage (disks) which can be used for running analyses such as converting a CRAM file to a BAM file or running a Jupyter Notebook to transform and visualize data.
1. Compute and disks - Concepts
GCE allows you to create and destroy VMs as you need them. You can create VMs of different shapes (CPU and memory) as you need them for different workloads.
GCE follows the cloud philosophy that you only pay for what you use in that you are only billed for VMs and disks between the time that you have created them and destroyed them. To be clear, however, you are "using" your CPU, memory, and disk space while your VM is running, even if your VM is sitting idle.
GCE's virtualization offers additional flexibility in that you can "stop" a running VM (at which point you stop being charged for the CPU and memory, but continue accruing charges for the disk) and "start" it again later. You can even change the amount of CPU and memory when you restart the VM.
In addition, GCE offers significantly reduced costs with preemptible VMs. If you have a workflow that will run in less than 24 hours, you can save up to 80% by using preemptible VMs.
2. GCE - Compute Costs
Detailing the GCE pricing flexibility is beyond the scope of this document. You are encouraged to get pricing details from the GCE Pricing documentation. In brief, your main questions to ask are:
- How many CPUs does my compute task require?
- How much memory does my compute task require?
- How much disk does my compute task require?
- Can my compute task finish in less than 24 hours?
If your compute need is for a long running compute node, then you should use a "full priced VM", since a preemptible VM lasts at most 24 hours. If your compute need is for less than 24 hours, and you can manage the complexity of preemption at any time within that 24 hours, a preemptible VM will cost almost 80% less. For more information, see the documentation on Preemption selection.
3. GCE - Disk Costs
GCE offers a range of disk types, including:
- Network attached magnetic disks (Persistent Disk Standard)
- Network attached solid state disks (Persistent Disk SSD)
- Locally attached solid state disks (Local SSD)
Pricing for the different disk types generally follows the rule that you pay more for large disks and more performant disks. Most life sciences workflows are not I/O bound and so the least expensive disk (Persistent Disk Standard) is typically the best choice. If your workflow is I/O bound, however, you may find that using Local SSDs on a preemptible instance is the best choice.
4. GCE - Data Egress
Egress charges apply when copying data out of the zone that a Compute Engine VM is running in. For example:
- Downloading data to your workstation or laptop
- Copying data from a VM in one zone to a VM in another zone
- Copying data from a VM in one region to a GCS bucket in another region
If data is copied between VMs in the same zone, no egress charges will be incurred. If data is copied between a VM and a GCS bucket in the same zone, no egress charges will be incurred. For example, accessing AMP PD data (stored in us-central1) using a Google Compute Engine VM running in us-central1-a will incur no egress charges.
Google BigQuery
Google BigQuery (BQ) is a database where "tables" are stored in "datasets". You can issue SQL queries to filter and retrieve data in BigQuery.
1. BigQuery Overview
As an example, AMP PD clinical data is stored in a dataset named amp-pd-research.2019_v1beta_0220, and demographics data for participants can be found at the path:
amp-pd-research.2019_v1beta_0220.Demographics
You can store both tabular data and nested data in BigQuery.
Storage has a cost associated with it. In addition when you query data in BigQuery, you want to consider just how much data your query "touches" as BigQuery query billing is based on the amount of data that the query engine "looks at" to satisfy the request.
2. BigQuery - Storage Costs
BigQuery storage costs are $0.02 per GB for the first 90 days after table creation and $0.01 per GB from then on.
3. BigQuery - Query Costs
When you run a query, you're charged according to the number of bytes processed in the columns you select or filter on, even if you set an explicit LIMIT on the number of records returned. This means that you want to be careful about which columns you put in your SELECT lists and WHERE clauses.
BigQuery query costs are $5.00 per TB with the first 1 TB per month free.
AMP PD clinical data is quite small. Each table is measured in KB or MB, not in GB or TB. You would need to query these tables very heavily to generate any meaningful charges.
AMP PD genomic variant data is quite large. The table amp-pd-research.2019_v1beta_0220_genomics.passing_variants is 2.11 TB. Thus a naive SELECT * FROM passing_variants query would cost $10.55.
BigQuery offers a number of features to help control query costs. See:
- BigQuery best practices: Controlling costs
- Estimating query costs
- Partitioned Tables
- Clustered Tables
In addition see the Jupyter notebook, Py3 - WGS - Query Variants, for insight into querying the AMP PD passing_variants table.
4. BigQuery - Egress Costs
BigQuery does not include explicit network egress charges, however BigQuery has limits on the amount of data that one can egress. A query has the limit:
- Maximum response size — 10 GB compressed
(When issuing a query that returns a large amount of data, you may write the results to another BigQuery table or to a GCS bucket.)
Cloud Use Cases
With the key Cloud costs listed above, we can revisit the original uses cases and provide the framework for their costs. Specific costs will vary based on software versions, data sizes, storage and access locations.
1. Running a Workflow
The Terra environment provides a workflow engine called Cromwell. The following discussion assumes that a workflow has been crafted to run using Cromwell. Many workflows in cloud, independent of the workflow engine, will follow a similar model.
A typical single-stage workflow follows the model:
- Create Compute Engine VM
- Copy inputs from GCS to the VM
- Run code on the VM
- Copy outputs from the VM to GCS
- Delete the VM
A typical multi-stage workflow is a sequence of single-stage workflows. A more complex multi-stage workflow may run intermediate stages in parallel. The core cost considerations for running all such workflows is the same:
- How much compute do you need (CPUs, memory, and disk)
- Can you run on preemptible VMs
- Can you run your compute nodes in the same region as your data in GCS (to avoid egress charges)
Whenever possible, run your VMs in the same region as your data. AMP PD data is stored in a GCS regional bucket in us-central1. If you run your VMs in a different region, you will incur network egress charges. If you run your VM in the same region as your data, you incur no network egress charges.
2. Converting a CRAM to a BAM
A typical 30x WGS CRAM file is about 17.5 GB in size. Let's look at the cost of converting a CRAM file to a BAM file.
For this example, we used a workflow published in Dockstore (seq-format-conversion/CRAM-to-BAM) and made available via the help-gatk/Seq-Format-Conversion workspace.
The workflow has two stages:
- CramToBamTask (Convert CRAM to BAM)
- ValidateSamFile (Validate the BAM)
Let's focus on CramToBamTask. Converting a CRAM to a BAM is a single threaded operation, and there is likely little to no advantage to allocating more CPUs. There could be advantages to adding more memory, but at certain memory sizes, GCE requires you to add more CPUs, which increases cost.
There is also cost for the persistent disk. For this operation, a 200 GB disk was allocated, which at $0.04 / GB / month is $8 per month or $0.01 per hour.
Let's look at a few configurations tested:
| Machine | Hourly Cost (preemptible/full price) | Runtime | Cost on preemptible VM | Cost on full priced VM |
|---|---|---|---|---|
| 4 CPUs 15GB (n1-standard-4) |
$0.04 / $0.19 | 4h 47m (4.62h) |
$0.051 * 4.62 = $0.24 | $0.20 * 4.62 = $0.92 |
| 1 CPU, 3.75 GB (n1-standard-1) |
$0.01 / $0.0475 | 5h 51m (5.85h) |
$0.02 * 5.85 = $0.12 | $0.0575 * 5.85 = $0.34 |
| 1 CPU, 6.5 GB (custom) |
$0.012 / $0.059 | 5h 26m (5.43h) |
$0.022 * 5.43 = $0.12 | $0.069 * 5.43 = $0.37 |
So the smallest VM shape had the highest runtime, but the least cost.
There are many things to highlight from this example:
- Preemptible VMs make a significant price difference.
- Between CPUs and memory, adding more memory is much less expensive.
- Adding more CPUs can decrease runtimes, but at a significant cost multiple.
- Increasing CPU or memory allocations can reduce the amount spent on disk (by shortening runtime), but disk is the least expensive of the three resource type
Note that at 17.5 GB, downloading the CRAM file to convert to BAM on your own workstation would cost $0.175.
While the total computational costs discussed in this example are all very small, let's be sure to look at what happens when you scale up the number of samples to 1000:
| Operation | Samples | Estimated Cost |
|---|---|---|
| CRAM to BAM 1 CPU, 3.75 GB, preemptible |
1000 | $120 |
| CRAM to BAM 4 CPUs, 15 GB, full price |
1000 | $920 |
| Download | 1000 | $175 |
3. Aligning a genomic sample to a reference and performing variant calling using the GATK
The Broad Institute has published the five-dollar-genome-analysis-pipeline to Dockstore and made it available in the help-gatk/five-dollar-genome-analysis-pipeline Workspace. Read through the workspace description for example costs of running the workflow.
Be aware that the "five dollar genome" is named for the typical amount of Compute Engine charges generated during processing of a 30x WGS sample. As important (if not more important) is the costs associated with file storage.
Long Term Storage
A typical 30x WGS sample produces a 17.5 GB CRAM file and a 6.5 GB gVCF file. Long term storage of these outputs in a Regional bucket ($0.02 / GB / month) would be:
| Samples | Monthly Cost | Annual Cost |
|---|---|---|
| 1 | $0.48 | $5.76 |
| 100 | $48.00 | $576.00 |
| 1000 | $480.00 | $5,760.00 |
Short Term Storage
Multi-stage workflows like the "five dollar genome" store inter-stage results in Google Cloud Storage. Such workflows store large interim results, such as complete BAM files or shards of FASTQ files. It is very important to clean up the interim results when you are done with the workflow. Inattention to cleaning up this storage can significantly increase your per-sample costs.
A typical single sample processing of a 30x WGS sample, can produce more than 300GB of interim storage. This is more than 12x the size of the final outputs. Storing these interim results for a month would cost (in a Multi-regional bucket @ $0.026 / GB / month):
| Samples | Monthly Cost |
|---|---|
| 1 | $7.80 |
| 100 | $780.00 |
| 1000 | $7,800.00 |
4. Aligning a transcriptomic sample to a reference using STAR
AMP PD Transcriptomics data was aligned using STAR version 2.6.1d. The workflow can be found at TODO.
Processing cost for most samples was between $1.50 and $2.50. As a single-stage workflow, there were no intermediate results to clean up. There are a few details to take away from this workflow:
- The per-sample cost was kept lower primarily by using preemptible VMs. Using a full priced VM would be more than 4 times as expensive.
- Having a separate workflow using samtools to sort and compress the BAM shortened total runtimes and allowed more samples to be processed with preemptible VMs.
- Having a separate workflow using samtools to sort allowed us to reduce the disk size from 1 TB down to 200 GB.
- Using a higher compression level (samtools defaults to 6, STAR defaults to 1) can save significantly on long term storage costs.
Compute Analysis
The STAR alignReads workflow used 16 threads (--runThreadN 16), and we used a VM defined as custom (16 vCPUs, 80 GB memory) and (initially) 1 TB of persistent disk. At preemptible rates, this VM is approximately $0.178; at full price, this VM is approximately $0.844/hr .
Preemptible VMs must finish their work within 24 hours. We observed that large samples and samples with high multi-mapping rates could take 24 hours or more. The impact of this is that a sample that could take 23 hours on a preemptible VM would cost $4.09 for compute, while a sample that took just over 24 hours on a full priced VM would cost $20.26 for compute.
To get more samples to complete in less than 24 hours, we changed the workflow such that STAR would not sort the BAM. This saved between 1-2 hours for the workflow.
This did mean that we needed to create another workflow to compress the BAM. However, we already had a need to generate a BAM index file, thus we allowed samtools to compress and re-index the BAM. We found that we could index BAMs on a small, single-core VM (n1-standard-1) with a 200 GB disk. At preemptible rates of $0.01 per hour for the VM and $0.01 for the disk, these workflows cost pennies to complete for each sample.
Storage Analysis
Note that the STAR default compression level is 1, while samtools default compression level is 6. Thus when we had samtools recompress the BAM files, we saw a 25% reduction in size. This has a tremendous long term cost benefit. An average sized RNASeq BAM file for AMP PD is 20 GB (level 6 compression) vs 27 GB (level 1 compression).
The cost of storing these BAMs in Regional storage would be:
| Number of BAMs | Level 1 compression (mo/yr) | Level 6 compression (mo/yr) |
|---|---|---|
| 1 | $0.54 / $6.48 | $0.40 / $4.80 |
| 100 | $54.00 / $640.80 | $40.00 / $480.00 |
| 1000 | $540.00 / $6,408.00 | $400.00 / $4,800.00 |
Running a Notebook
The Terra environment provides the ability to run analyses using Jupyter notebooks. In this section, we look at costs around using the Jupyter notebook service, along with costs for running a couple of example notebooks.
1. Notebooks Section Overview
For fully understanding the notebook environment, you are encouraged to read these articles:
- Terra's Jupyter Notebooks Environment Part I - Key Components
- Terra's Jupyter Notebooks Environment Part II - Key Operations
When breaking down the costs for using the notebook service, there are two broad categories to look at:
- Compute
- Egress and Query costs
2. Notebooks - Compute Costs
Your compute costs are based on the VM that is allocated for you , whether that VM is doing any computation or not. Consider:
- A GCE VM is created for you when you open your first notebook for editing.
- While the VM is running, you will be charged for the allocated CPUs, memory, and disk.
- You can pause the VM
- While the VM is paused, you will only be charged for the disk.
- You can delete the VM
- When the VM is deleted, you will not be charged.
Example runtime costs
By default, your Notebook Runtime allocates a VM with 4 cores, 15 GB of memory (n1-standard-4), and a 500 GB disk.
The cost for this VM and disk are:
VM: $0.190 / hour
Disk: $0.027 / hour (approximate)
So while the VM is running, you will be charged about $0.217 per hour. When your VM is paused, you will be charged $0.027 / hour.
If you were to have a notebook VM running for 20 hours per week, your weekly charges would be:
($0.217 / hr * 20 hr) + ($0.027 / hr * 148 hr) = $8.34
or monthly costs of about $33.34.
3. Notebooks - Egress and Query costs
Whether your notebook is running or sitting idle, you incur the same compute charges. How your notebook access available data determines any additional charges. For this section, we look at whether your data is in Google Cloud Storage or in BigQuery to assess any additional charges.
Google Cloud Storage
If your data is in GCS and is in the same region as your notebook VM, then you pay no access charges. As of the time of writing, Terra notebook VMs run in zones in us-central1. AMP PD published data is in us-central1. Thus no egress charges are incurred.
Google BigQuery
If your data is in BigQuery and the data is "small", then you are likely to incur no additional costs for accessing the data. BigQuery query pricing is "$5.00 per TB" and "First 1 TB per month is free".
All of the AMP PD clinical data is less than 100 MB, thus you would have to query all of the AMP PD clinical data more than 10,000 times in a single month before you would incur charges. After that, you would have to query all of the AMP PD clinical data 200 times before you are charged $0.01.
If your data is in BigQuery and the data is "large", then you will want to pay close attention to how you query the data. Review the discussion above on BigQuery Query costs.
Example notebooks
Performing Quality Control checks on genomic data
A notebook that performs quality control checks on genomic is typically driven by "analysis ready" data, such as QC metrics emitted by the Picard set of tools. In such cases, if the metrics are aggregated in BigQuery (as they are for AMP PD), the data is very small (on the order of MB) and thus is virtually free to query.
If the notebook also queries a large table such as a table of variants, you may begin to generate notable charges. A query that selects all of the values from AMP PD passing_variants table will cost over $10, while a more compact and targeted query can be much less.
Analyzing genomic variants
If you are looking at a targeted region of the genome, querying the AMP PD passing_variants table can be fairly inexpensive. The passing_variants table takes advantage of BigQuery clustering. The table is clustered on the reference_name (the chromosome), the start_position, and the end_position. What this means is that when you know the region of interest you can direct BigQuery to just look at the cluster where our data of interest is.
For example, a query that includes in the WHERE clause:
reference_name = 'chr4'
will look only at the records for chromosome 4 (which is less than 7% of the genome), and so the cost of the query will be less than 7% than if you queried the entire table.
If you are looking at the entire genome with your analysis, be mindful of your queries as you can generate meaningful charges. It may be worth evaluating other options which include processing VCF files on a VM. Copying the VCF(s) from GCS eliminate the query charges. This is not always the better solution, as you may only be trading off query time for increased compute time. These types of trade-offs require deeper analysis of the specific use case.
4. Storing Files in Cloud
In this section, we provide a quick look at what it costs to store certain types of data, on average. For your own data, you are encouraged to use the Cloud Storage Pricing Guide.
Notebooks
Notebook files typically store very little data, and notebook file contents (code and text) is quite small. An analysis of Jupyter notebooks in github indicates an average size of 600 KB. At $0.026 per GB per month (multi-regional), 1000 notebooks (600 MB) will cost about $0.016 per month.
Clinical Data
Clinical data is typically small. AMP PD clinical data in GCS is 19 MB. At $0.02 per GB per month (regional), this data costs $0.00038 per month.
Genomic Data
Genomic data is large. A typical 30x whole genome will have files of size:
- FASTQs: 75 GB
- CRAM: 17.5 GB
- gVCF: 6.5 GB
Depending on your access patterns, you should consider storing these files in Regional ($0.02 / GB / month) or Nearline ($0.01 / GB / month) storage.
FASTQ
| Samples | Monthly Cost (Regional) | Annual Cost (Regional) | Monthly Cost (Nearline) | Annual Cost (Nearline) |
|---|---|---|---|---|
| 1 | $1.50 | $18.00 | $0.75 | $9.00 |
| 100 | $150.00 | $1,800.00 | $750.00 | $900.00 |
| 1000 | $1,500.00 | $18,000.00 | $7,500.00 | $9,000.00 |
CRAM
| Samples | Monthly Cost (Regional) | Annual Cost (Regional) | Monthly Cost (Nearline) | Annual Cost (Nearline) |
|---|---|---|---|---|
| 1 | $0.35 | $4.20 | $0.175 | $2.10 |
| 100 | $35.00 | $420.00 | $17.50 | $210.00 |
| 1000 | $350.00 | $4,200.00 | $175.00 | $2,100.00 |
gVCF
| Samples | Monthly Cost (Regional) | Annual Cost (Regional) | Monthly Cost (Nearline) | Annual Cost (Nearline) |
|---|---|---|---|---|
| 1 | $0.13 | $1.56 | $0.065 | $0.78 |
| 100 | $13.00 | $156.00 | $6.50 | $78.00 |
| 1000 | $130.00 | $1,560.00 | $65.00 | $780.00 |
5. Transcriptomics Data
Transcriptomic data is large. A typical 100 million read RNA seq will have FASTQs and BAMs, each around 15 GB. Depending on your access patterns, you should consider storing these files in Regional ($0.02 / GB / month) or Nearline ($0.01 / GB / month) storage.
Storing either of these will cost approximately:
| Samples | Monthly Cost (Regional) | Annual Cost (Regional) | Monthly Cost (Nearline) | Annual Cost (Nearline) |
|---|---|---|---|---|
| 1 | $0.30 | $3.60 | $0.15 | $1.80 |
| 100 | $30.00 | $360.00 | $15.00 | $180.00 |
| 1000 | $300.00 | $3,600.00 | $150.00 | $1,800.00 |
General Advice
The following is general advice for controlling costs when using Google Cloud for typical life sciences work. The following "quick tips" are explained in more detail below:
1. Storage Best Practices
While many life sciences projects will commit a lot of time and energy into optimizing their data processing workflows, it is often long term storage costs that will dominate the budget. The reason for the high storage costs is the large data that is generated in the life sciences, such as genomic and transcriptomic. The following sections provide tips for keeping storage costs of large data under control.
Use Regional storage
For life sciences data, there is rarely a reason to make the data available in multiple Google Cloud regions. The cost of Regional storage is 77% of Multi-regional storage. The easiest way to save your project 23% is to put your data and compute in a single region. AMP PD data is stored in us-central1.
Compress large data
Compression rates vary, but some common options are:
- Compress STAR-generated BAMs (and index them) with samtools (discussed above)
- Convert WGS BAMs to CRAMs (and index them) with samtools
- Compress VCFs with bgzip (and index them with tabix)
Move data to cold storage (Nearline or Coldline)
Determining whether you can move large files to cold storage can tricky. If you move files that are accessed frequently, the access charges can wipe away the storage savings. However, much life science data goes through a life cycle of:
- Source data is generated
- Source data is processed into smaller summary information
- Summary information is used extensively
- Source data is used rarely
FASTQ files for genomics and transcriptomics fit this model and are large. Moving these files to Nearline after initial processing can save a project a lot of money on its largest data.
Clean up workflow intermediate files promptly
WDL-based workflows on large files, such as FASTQs, BAMs, and gVCF often have intermediate stages where large files are sharded or converted to different formats, creating many artifacts that get stored in Google Cloud Storage. Leaving these files in Cloud Storage can result in significant costs associated with running workflows. If the workflow succeeds, clean up the intermediate files, especially the large ones.
2. Compute Best Practices
Many people in the life sciences are familiar with working in an HPC environment. In this case, they have a compute cluster available to them. This cluster is typically a modest fixed size and is often shared with other researchers and departments. Thus the primary driver for computation is toward having jobs finish quickly and to minimize compute resources (CPUs, memory, and disk).
In this environment, if you have 1,000 samples to process, each takes a day to process, and available computing for 100 samples to run concurrently, then such processing will finish in 10 days (if all goes well). If you can reduce the time to process a single sample by 30%, you'll finish your processing in a week.
With cloud computing, you are generally not resource constrained in the same way. If you want to run 1,000 samples concurrently, you can generally do that (just be sure to request more Compute Engine Quota; if working in Terra, see this article). Reducing runtimes and compute resources will save you money, but you have other knobs to turn on money saving notably with preemptible VMs. Life science workflow runners, like Cromwell, are designed to take advantage of preemptible VMs).
As a general approach to saving on compute costs, approach optimization in the following order:
- Use preemptible VMs
- Reduce the number of CPUs (they are the most expensive resource)
- Reduce the amount of memory (add monitoring to your workflows)
- Reduce the amount of disk used (add monitoring to your workflows)
Below are some specific suggestions around preemptible VMs and monitoring.
Preemptible VMs
Cromwell has the ability to use preemptible VMs and for each task, you can set a number of automatic retries before falling back to a full priced VM. Some additional details to know about using preemptible VMs:
- Smaller VMs are less likely to be preempted than large VMs
- Preemption rates are lower during nights and weekends
- IO-bound workflows may benefit from using Local SSDs on preemptible instances
- Preemptions tend to happen early in a VMs lifetime
This last bullet point is important to understand. It is explained further in Google's documentation:
Generally, Compute Engine avoids preempting too many instances from a single customer and will preempt instances that were launched most recently. This might be a bit frustrating at first, but in the long run, this strategy helps minimize lost work across your cluster. Compute Engine does not charge you for instances if they are preempted in the first minute after they start running.
So while running on a preemptible VM and getting preempted adds cost overhead (cutting into your savings), such preemptions tend to happen early and the additional cost is modest.
Monitoring
It is difficult to save on CPUs, memory, and disk if you don't know your peak usage while workflows are running. Adding a little bit of monitoring can go a long way. Observations you may make about a workflow stage, once you have added monitoring:
- This workflow stage for the largest sample uses
- about the same <cpu, memory, disk> as the smallest sample
- much more <cpu, memory, disk> as the smallest sample
With this information, you can decide whether it is worthwhile to adjust <cpu, memory, disk> on a per-sample basis.
You might also observe:
- This workflow stage runs a sequence of commands and the disk usage never goes down
- If we cleaned up intermediate files while running, we could allocate less disk for each workflow
- This workflow stage runs a sequence of commands; some are multi-threaded and take advantage of more CPUs and some commands are single-threaded.
- If we made this a multi-stage workflow, we could use a single CPU VM for some steps and reduce total CPU cost
- This workflow runs on an n1-standard-<n> machine, but we never use all of the memory, we could change to an n1-highcpu-<n> machine (or a custom VM).
3. Egress Best Practices
Moving data from a multi-regional bucket to a regional bucket incurs egress charges at a rate of $0.01/GB. This means, for example, that moving 100 TB of data from a Terra workspace bucket (multi-regional US) to your own Regional bucket will cost $1,000.
Suppose that 100 TB of data is made up of one thousand 100 GB files. You could create a workflow on Terra that runs 1000 concurrent n1-standard-1 preemptible VMs, each with a 200 GB disk to:
- Copy file from multi-regional bucket to VM
- Copy file from VM to Regional bucket
- Remove the file from the multi-regional bucket
Each VM + disk would cost approximately $0.02 per hour and would finish in less than 1 hour. Your cost for transfer is thus on the order of $20.