Unified AMP PD Cohorts
The foundation of AMP PD is predicated on Parkinson’s disease (PD) related study data that is collected, harmonized, and made accessible for identifying new biomarkers and developing new treatments for PD. Overcoming challenges associated with resources, time and cost, and data availability is made possible through the collective efforts of the Michael J. Fox Foundation (MJFF) and National Institutes of Neurological Disorders and Stroke (NINDS) BioFIND study, Harvard Biomarkers Study (HBS), the National Institute on Aging (NIA) International Lewy Body Dementia Genetics Consortium Genome Sequencing in Lewy body dementia case-control cohort (LBD), the MJFF LRRK2 Cohort Consortium (LCC), the NINDS Parkinson's disease Biomarkers Program (PDBP), MJFF Parkinson’s Progression Markers Initiative (PPMI), and the NINDS Study of Isradipine as a Disease Modifying Agent in Subjects With Early Parkinson Disease, Phase 3 (STEADY-PD3), and the Study of Urate Elevation in Parkinson’s Disease, Phase 3 (SURE-PD3).
harmonized, and made accessible for identifying new biomarkers and developing new treatments for PD. Overcoming challenges associated with resources, time and cost, and data availability is made possible through the collective efforts of the Michael J. Fox Foundation (MJFF) and National Institutes of Neurological Disorders and Stroke (NINDS) BioFIND study, Harvard Biomarkers Study (HBS), the National Institute on Aging (NIA) International Lewy Body Dementia Genetics Consortium Genome Sequencing in Lewy body dementia case-control cohort (LBD), the MJFF LRRK2 Cohort Consortium (LCC), the NINDS Parkinson's disease Biomarkers Program (PDBP), MJFF Parkinson’s Progression Markers Initiative (PPMI), and the NINDS Study of Isradipine as a Disease Modifying Agent in Subjects With Early Parkinson Disease, Phase 3 (STEADY-PD3), and the Study of Urate Elevation in Parkinson’s Disease, Phase 3 (SURE-PD3).
When unified, data from these eight cohorts offer the PD research community unprecedented access to a large, harmonized dataset with common clinical and genomic data, bridging the gap between prohibitive individual research constraints with large-scale, state-of-the-art analysis.
Introduction to the Cohorts
Cohort Data & Genetic Criteria
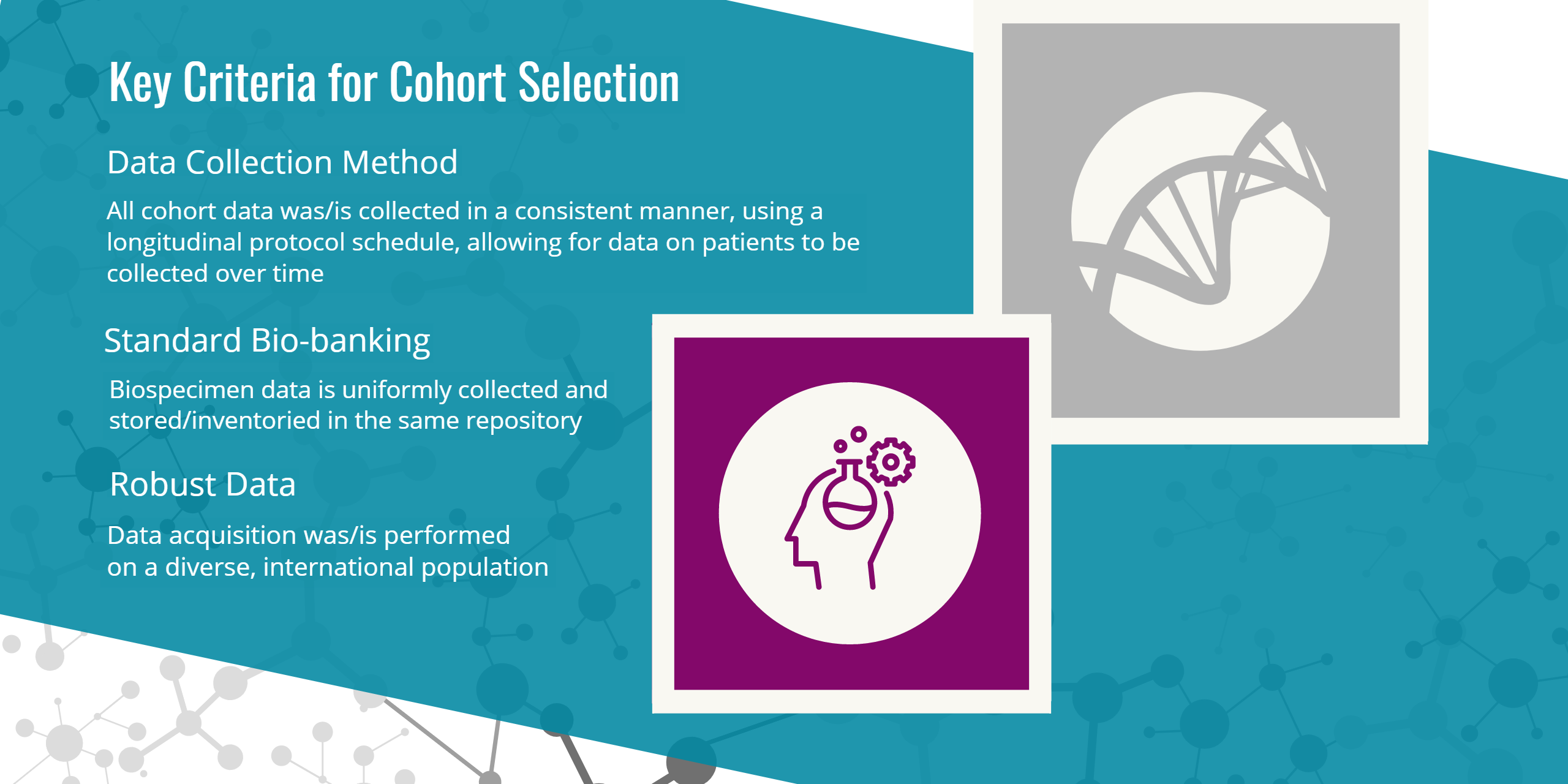
All AMP PD Cohorts provided clinical data and at least one set of DNA, RNA, and Proteomic assay data. The following criteria was used to assess each of the four Cohorts and provides additional details for why each was selected and found suitable for inclusion:
- Relevance to PD research
- Dataset size (cases and controls)
- Patient consent includes approval for broad data-sharing of de-identified clinical and molecular data
- Adherence to the AMP PD DUC
- Longitudinal study method
- Uniform and standardized clinical data collection instruments
- Biosample availability with quality control metrics
- Inclusive of pathological sample data
- Cost-benefit analysis regarding storage, data harmonization, and analytical costs is in the interest of the AMP PD community