News & Updates
New Polygenic Risk Scores Workspace Available!
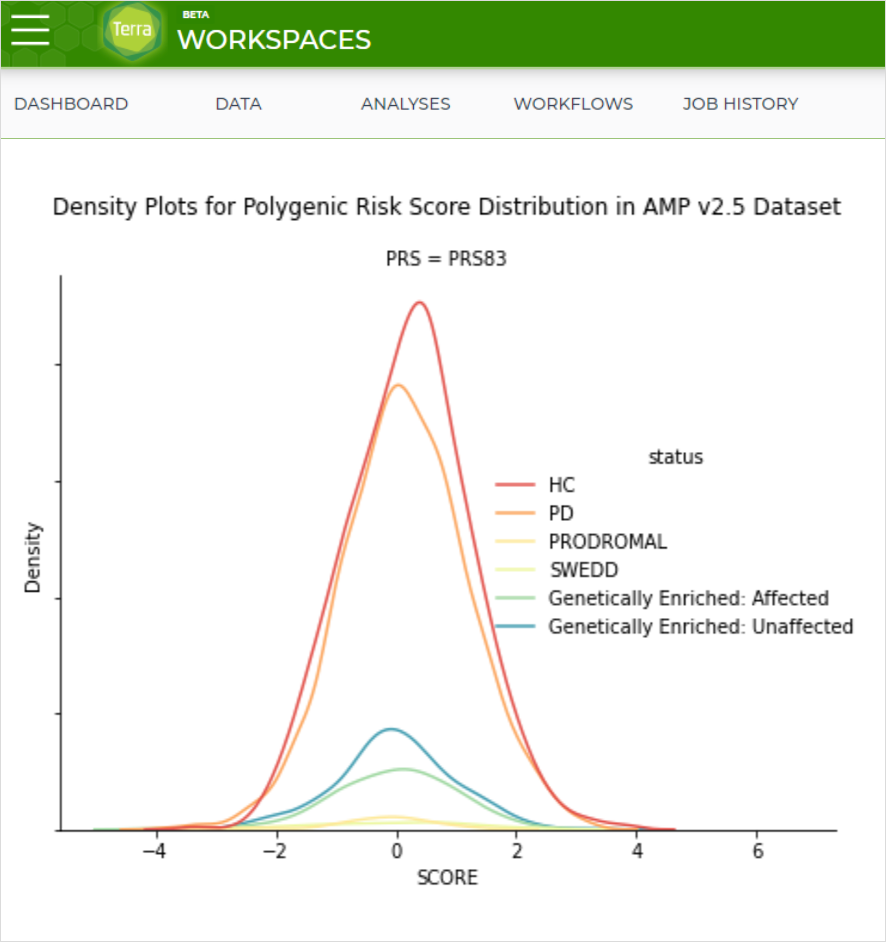
AMP PD has released a new workspace in Terra, utilizing AMP PD clinical and genetic data to calculate polygenic risk scores for AMP PD participants.
Parkinson’s disease is a complex neurodegenerative disorder believed to be caused by a combination of genetic and non-genetic factors. Recent advances in genome-wide associate stories (GWAS) have enabled the identification of several genetic loci that collectively contribute to PD risk. Exploring known genetic loci can shed light on this complicated issue and provide new ways to explore genetic risk for an individual or group of individuals.
What is PRS?
A polygenic risk score (PRS) is used to evaluate one's overall risk or predisposition to a disease, taking into account an individual’s genetic architecture that includes the effects of both protective and risk genetic variants.
This value is calculated by comparing individual-level genotype and phenotype data to a larger collection of weighted alleles from genome-wide association study summary (GWAS) statistics and this value indicates an individual's susceptibility based on alleles present in their genome. It is important to note that PRS scoring represents only one form of PD risk, and that the many variants used in assigning a PRS score require careful evaluation in the course of analysis.
How is PRS Useful?
- Calculating PRS can differentiate cases from controls on a population or group level: Cases are expected to have a significantly higher mean PRS than controls
- Can advise exploration on different pathways or biomarkers: PRS summarizes many genetic variants associated with PD risk. It is expected that a true biomarker of risk or disease will more likely be present in persons with a high PRS than with a low PRS. The specific SNPs which collectively make up a PRS may help identify mechanisms which are disrupted in PD to identify biomarkers or therapeutic targets.
- Stratify Parkinson’s Disease patients based on genetic risk: Linear PRS can be calculated based on GWAS SNPs enriched in given cellular pathways. This allows comparison of different risk groups or potential mechanistic subtypes to better understand heterogeneity of disease and the role of specific genetic variants in the clinical features of patients and controls.
- Don’t need a large sample size: PRS relies on the cumulative risk from many genomic loci and can be calculated for any number of individuals and get meaningful results.
Within this workspace, users are able to generate covariate files based on AMP PD features that have been captured and harmonized across cohorts, and use these to determine polygenic risk scores, which can then evaluate a participant’s risk or predisposition for PD. This workspace provides a way to conveniently perform this across AMP-PD cohorts consistently to explore, discover, and analyze the rich data present within the platform.
To find this workspace and more, please see the workspace directory in our AMP PD featured workspace here.