News & Updates
AMP PD v3 Release Notes - December 2022
AMP PD is pleased to announce to the community that the next release of data has been made to the AMP PD Knowledge Portal and AMP PD in Terra! All users who have currently valid DUA agreement will be able to access the data in the portal.
Data Summary
Data Composition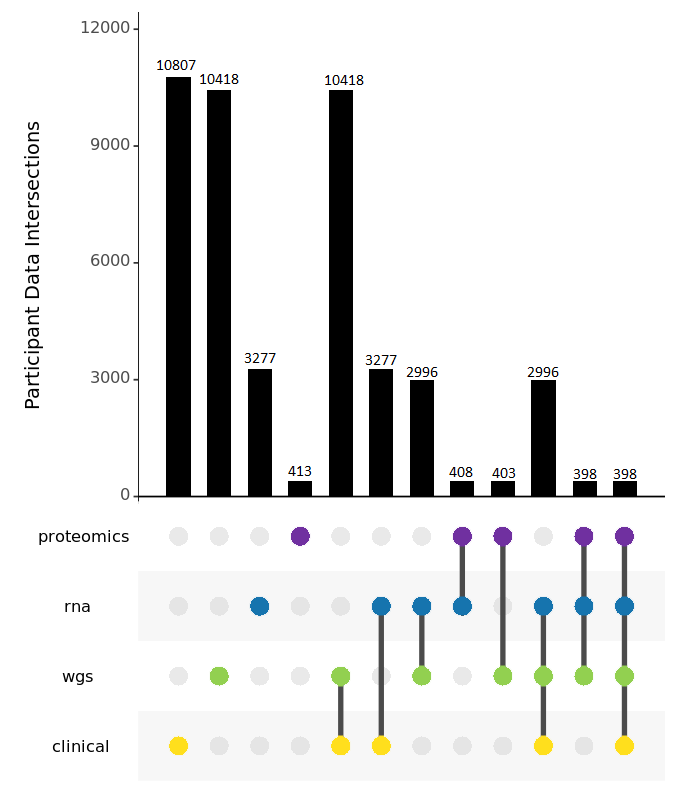
AMP PD's release 3.0 public dataset includes 10,807 participants from eight cohorts, including BioFIND, HBS, LBD, LCC, PDBP, PPMI, Steady PD, and Sure PD. AMP PD includes clinical data for all subjects, as well as whole genome sequencing data for 10,418 joint genotyped participants, transcriptomics data for 3,277 participants, and targeted proteomics data for 413 participants.
This release includes 2996 subjects with fully integrated clinical records, WGS joint genotyped samples, and RNA samples and, for 398 of these subjects, includes targeted proteomics data as well.
Release 3.0 Data
The data products within Release 3.0 include targeted proteomics data generated on the Olink platform by both Olink and AbbVie, Whole Genome Sequencing (WGS) data from 57 samples not included in previous releases, Parkinson’s Progression Markers Initiative (PPMI) GUID updates, Plink 1.9 data updates, Lewy Body Dementia cohort WGS gap metrics data (i.e., missing metrics files), and clinical data (including demographics data) updates.
As with past data releases for AMP PD, the data for Release 3.0 is stored in Google Cloud Storage (GCS) and Google BigQuery structures. The proteomics data are stored in files named based on tissue source, assay type, and file version and are stored in BigQuery within four different datasets: Olink CSF data, AbbVie CSF data, Olink plasma data, and AbbVie plasma data. For each of these four datasets, four types of data tables are available: cardiometabolic, inflammation, neurology, and oncology. In GCS, these data tables are stored as matrix files, and Olink Explore files are also accessible for researchers using Olink-specific tools. If researchers have questions on these new data buckets and BigQuery tables, they can contact the AMP PD team at admin@amp-pd.org.
The metadata tables generated for Release 3.0 as with past releases will store participant IDs, sample IDs, visit months, data product codes, tissue sources, assay types, assay codes, file version definitions, and file version codes. The metadata tables will also include matrix file and Olink Explore file links to the cardiometabolic, inflammation, neurology, and oncology data tables.
In addition, the team has assembled Getting Started notebook updates (e.g., filtering sample duplicates). The new and updated workspaces for Release 3.0 can be found at the following links:
- Getting Started Tier 1 - Clinical Access
- Getting Started Tier 2 - Clinical and Omics Access (must be logged in to Tier 2 access link)
- AMP PD - Proteomics QC and Analysis (must be logged in to Tier 2 access link)
In addition, they can be found in the Workspace Showcase and the Dataset Library.
Global Inventory Table
As another part to this latest release, the team has created a global inventory table to help investigators better understand what data is available for each patient within the AMP PD data. To avoid the need for complex queries against different files, the AMP PD team developed the new Global Inventory table, which can be searched using participant ID and visit month information. Table headings list the different available data products and table cells contain sample names. This table provides users with a method to search the entire AMP PD data inventory and allows users to compare the data products released in Release 3.0 with previous releases.
V3.0 Release Updates
Additions
- New data type: Official Release of 2 Targeted Proteomics Datasets
- New WGS Single Sample Data
- New GUID values for PPMI participants
- New Terra workspace: AMP PD - Proteomics QC and Analysis
- New Global Inventory Metadata Table