News & Updates
AMP PD Release 4.0 Release Notes – November 2023
AMP PD is pleased to announce to the community that the Version 4 data release is now available through the AMP PD Knowledge Portal and AMP PD in Terra! All users who have a currently valid DUA agreement will be able to access the new data in the portal.
New in this release:
- Postmortem Brain Cohort from 100 Single Nucleus participants with clinical data, WGS, and RNASeq data with samples from 5 brain regions, with single-sample and aggregate sequence, bam, vcf, and h5ad data
- Untargeted Proteomics Data from 621 participants from PDBP and PPMI cohorts, with 4278 samples from plasma and CSF sources at the fragment, protein, and peptide levels, along with normalized batch corrected matrix data
- Targeted Proteomics Data has been normalized from two datasets comprising 413 PDBP and PPMI participants in 5 targeted CSF and Plasma assays to enable analysis across all previously released targeted proteomics data
Data Composition
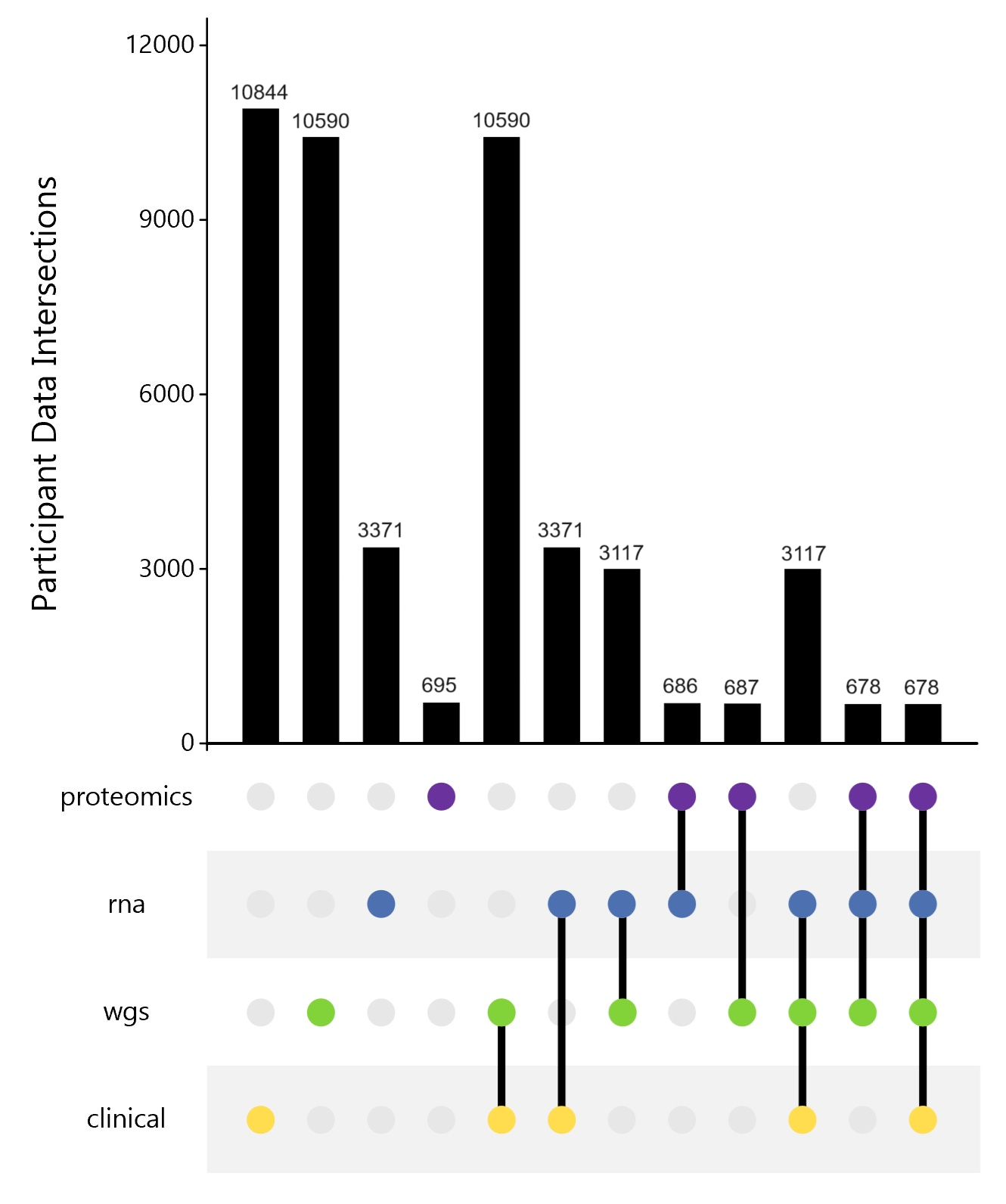
AMP PD's release 4.0 public dataset includes 10,908 participants in a Unified Cohort comprising nine cohorts that include BioFIND, HBS, LBD, LCC, PDBP, PPMI, Steady PD, Sure PD and the new Postmortem Cohort.
AMP PD includes clinical data for all participants, as well as WGS data for 10,418 joint genotyped samples, transcriptomics data for 3,364 participants that includes 8461 whole blood bulk RNA samples, proteomics data from 695 participants in targeted and untargeted proteomics datasets, and single nucleus data from 100 participants with matching clinical, WGS, and RNA samples from 5 brain regions.
Within this release, 3007 participants have matching clinical records, WGS joint genotyped samples, and RNA samples. Of these participants, 672 participants also have overlapping proteomics data within targeted and untargeted datasets.
Release 4.0 Data
Proteomics
Proteomics within Release 4.0 includes a bridged targeted proteomics dataset that was generated using the Olink platform from CSF and Plasma targeted assays in 5 panels, and untargeted mass spectrometry based proteomics data from plasma and CSF sources at the fragment, protein, and peptide levels, along with normalized batch corrected matrix data.
Targeted Proteomics: The bridged targeted proteomics datasets comprise CSF and plasma based assays in 5 panels: cardiometabolic, inflammation, neurology, and oncology. These are released in file based formats and as queryable tables in BigQuery. NPX files for each of CSF (CSF) and Plasma (PLA) tissue sources are available in per-panel files and aggregated matrix files. Per-panel files have also been released in Explore format for use with OLink analysis tools.
Four types of data tables for each of CSF (CSF) and Plasma (PLA) tissue sources are available in BigQuery and can be accessed directly through Google interfaces or through notebooks and tools in Terra. AMP PD provides examples for how to access, retrieve, and use the targeted proteomics data through workspaces and notebooks in Terra. Registered users can access the workspaces below, which include Jupyter notebooks written in R and in python that can be cloned and edited to suit your needs
AMP PD - Proteomics Getting Started
The purpose of this workspace is to provide information to help you get started. This workspace includes directory listings and notebooks to help access, retrieve, analyze, and download proteomics data
AMP PD - Proteomics QC and Analysis : Release 4.0
The purpose of this workspace is to provide QC and analysis notebooks for AMP PD Tier 2 users.
- Targeted Proteomics PCA: graphing principal component analysis
- NPX Boxplots: subset proteomics data to generate boxplots
- APOE Proteomics Case Study: protein expression and APOE genotype correlation, case/control deltas
- QCPlots: simple quality control checks and QC plots to show a visual representation of the QC data
Untargeted Proteomics: The untargeted proteomics datasets comprise CSF and plasma based assays that use mass spectrometry via the Orbitrap Exploris platform and Openswath target extraction software to produce raw, processed, and aggregate expression data at the fragment, protein, and peptide levels. Single sample data is available for the CSF (CSF) assay in raw and mzML formats, along with normalized batch corrected matrix data in csv formats. The plasma (PLA) assay includes native and depleted single sample and aggregate data in the same file formats. And each of CSF and PLA datasets is accompanied by a complete SDRF metadata file that includes relevant clinical data fields and instrument details.
AMP PD strives to provide clear provenance of the data. As such, this release contains files at each stage of processing to allow researchers to see how the data evolved and to allow researchers to process samples in a divergent manner. Intermediate data files in this release include matrices that are prepared before batch correction was applied, before imputation was applied, and before normalization was applied at the fragment level.
A data table for each of CSF (CSF) and Plasma (PLA) assays identifies protein abundance per detected UniProt id for each participant sample. These are available in BigQuery and can be accessed directly through Google interfaces or through notebooks and tools in Terra. AMP PD provides examples for how to access, retrieve, and use the untargeted proteomics data through workspaces and notebooks in Terra. Registered users can access the workspaces below, which include Jupyter notebooks written in R and in python that can be cloned and edited to suit your needs.
AMP PD - Proteomics Getting Started Tier-2
The purpose of this workspace is to provide information to help you get started. This workspace includes directory listings and notebooks to help access, retrieve, analyze, and download proteomics data
AMP PD - Proteomics QC and Analysis
The purpose of this workspace is to provide QC and analysis notebooks for AMP PD Tier 2 users.
- Untargeted Proteomics PCA: graphing principal component analysis
- NPX Boxplots: subset proteomics data to generate boxplots
- APOE Proteomics Case Study: protein expression and APOE genotype correlation, case/control deltas
Single Nucleus Brain Sample Data
With the introduction of Single Nucleus data, a new Postmortem cohort has been added to the AMP PD Unified Cohort. This data includes clinical, genomic, and transcriptomic data from single nucleus brain samples. Brain tissue samples from PD case and control participants were prepared for 5 brain regions: the primary visual cortex, primary motor cortex, prefrontal cortex, dorsal motor nucleus of the Xth nerve, and globus pallidus interna. Sample data available in this release includes single sample WGS data in cram and vcf formats, and single sample and aggregate RNA sequence data in h5ad.
BigQuery datasets for Single Nucleus WGS and RNA sequence data include AMP PD standard sample inventory data and metadata, which can be queried to learn file locations and participant data, and a QC flags data table. AMP PD provides examples for how to access and retrieve clinical, WGS, and RNA sequence data in the Single Nucleus cohort through workspaces and notebooks in Terra. Registered users can access the workspace below, which includes Jupyter notebooks written in R and in python that can be cloned and edited to suit your needs.
AMP PD - Getting Started Tier-2 - Clinical and Omics Access
The purpose of this workspace is to provide information to help you get started. This workspace includes directory listings and notebooks to help access, retrieve, analyze, and download proteomics data
- Clinical - load a table from BigQuery: interacting with and displaying clinical data in a Jupyter notebook
- Single Nucleus RNAseq - load H5AD from cloud storage: identify case and control participants and retrieve associated SN data
V4.0 Release Updates
Additions
- New cohort: Postmortem Brain Cohort with clinical WGS and RNA Seq data
- New data type: Untargeted Proteomics from mass spectrometry
- New dataset: Targeted Bridged dataset consolidating 2 previously released targeted datasets for cross analysis
- Recovered WGS: WGS sample that aligns with new proteomics data was recovered by meeting AMP PD clinical data requirements
- Terra Workspace Updates: Getting Started with Proteomics Notebook examples to use the Bridged Targeted Proteomics dataset
- Terra Workspace Notebooks: Getting Started Notebooks for using and analyzing Untargeted Proteomics
Changes
- Corrected BAM index files for EV exRNA pilot samples and recovered two BAM files