News & Updates
AMP PD Release Notes - October 2019
Data Summary
Data Composition
Clinical Data
Participant records were compiled from BioFIND, HBS, PDBP, and PPMI cohorts into a harmonized dataset. These records were then paired with RNA and WGS samples and excluded if matching sample data was not 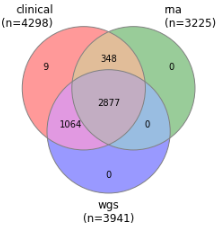 available, with the exception of 9 participants whose WGS samples were excluded solely for duplicating samples in AMP PD cohorts.
available, with the exception of 9 participants whose WGS samples were excluded solely for duplicating samples in AMP PD cohorts.
RNA Data
RNA sample data was sequenced and processed for BioFIND, PDBP, and PPMI cohort participants. RNA samples were excluded during QC rounds when there was no corresponding clinical data.
WGS Data
DNA sample data was sequenced and processed for BioFIND, HBS, PDBP, and PPMI cohort participants. WGS samples were not excluded during QC rounds when there was no corresponding clinical data or when available clinical records require further investigation to warrant sample exclusion. In the 2019_v1release_1015, all HBS samples were excluded from the joint genotyping run to address missing data sharing consents in one or more subjects, to be included in the next joint genotyping run once HBS participant consents are fully reconciled.
Integrated Data
This release includes 2877 subjects with fully integrated clinical records, WGS samples, and RNA samples. For an additional 348 participants, this release includes RNA samples with corresponding clinical records when WGS is not yet available. There are similarly 1064 WGS samples with clinical records where RNA sample data is not yet available. There are no cases where only RNA and WGS data intersect because RNA QC required that clinical records exist.
Composition by Cohort
BioFIND Data
Of 213 participants whose clinical records met AMP PD minimum clinical data criteria, 167 participants have corresponding samples in all three release data categories.
HBS Data
Of 876 participants whose clinical records met AMP PD minimum clinical data criteria, all 876 have corresponding WGS sample data. HBS is not represented in the joint genotyping data due to reconciliation of participant consents. HBS samples are not fully integrated as no RNA sequence data was processed for this release.
PDBP Data
Of 1599 participants whose clinical records met AMP PD minimum clinical data criteria, 1315 participants have corresponding samples in all three release data categories, 1469 have corresponding WGS samples, and 1445 have corresponding RNA samples.
PPMI Data
Of 1610 participants whose clinical records met AMP PD minimum clinical data criteria, 1395 participants have corresponding samples in all three release data categories, 1433 have corresponding WGS samples, and 1572 have corresponding RNA samples.