News & Updates
AMP PD Release Notes – May 2021
Data Summary
Data Composition
Clinical Data
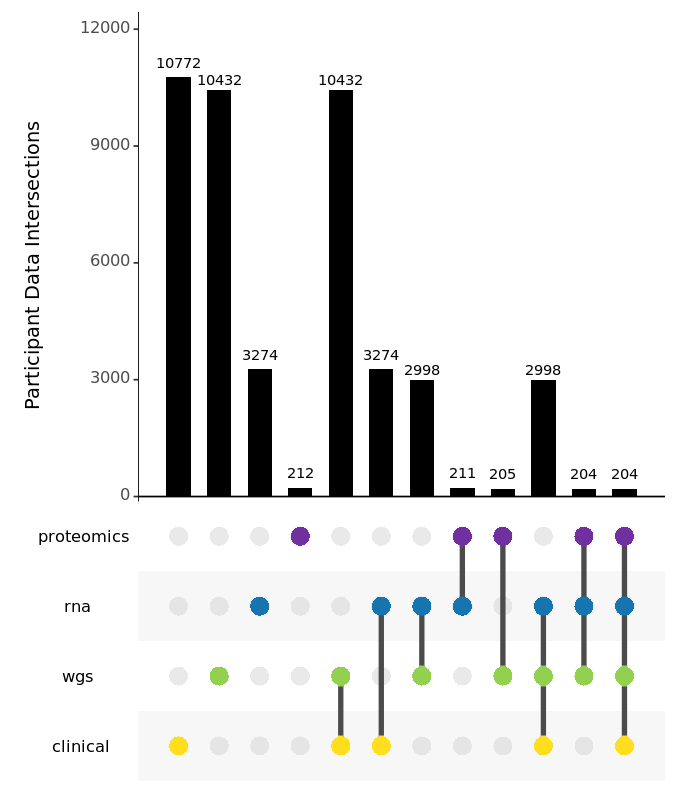
Participant records were compiled from seven cohorts and harmonized to form a single unified AMP PD cohort dataset. These records were then paired with RNA and WGS samples and excluded if matching sample data was not available, with 64 participants who appeared in multiple studies included by reference to unique participant samples. This release features a Preview of the new Targeted Proteomics dataset of 743 samples for 212 participants, all paired with clinical data and 204 of which have a complete set of matching WGS and RNA samples. Adding 531 WGS samples, this v2.5 release completes the Steady PD cohort, adds the new Sure PD cohort in full, and pushes the Joint Genotyped samples for the unified AMP PD cohort to 10,418 samples!
Integrated Data
This release includes 2998 subjects with fully integrated clinical records, WGS samples, and RNA samples. For an additional 276 participants, this release includes RNA samples with corresponding clinical records where WGS is not available. Similarly, there are 7434 WGS samples with clinical records where RNA sample data is not available.
RNA Data
RNA sample data was sequenced and processed for BioFIND, PDBP, and PPMI cohort participants. The AMP PD v2 release featured 8461 RNA samples for 3274 Participants. In the latest v2.5 release, the complete set of 192 associated RNA pooled samples have been added, along with the same set of artifacts AMP PD has previously released with the single sample data. This includes products from Salmon, STAR, Featurecounts, Picard metrics, and aggregated summary reports as files in Google Cloud Storage and as queryable tables in BigQuery.
WGS Data
DNA samples were sequenced and processed through the Broad's GATK pipeline for BioFIND, HBS, LBD, LCC, PDBP, PPMI, Steady PD, and Sure PD cohort participants. WGS samples were excluded from the v2.5 release when there was no corresponding clinical data. All WGS samples were vetted through a series of independent genomic QC checks and interdependent multi-modal QC checks.
In the v2.5 release dataset, 532 new samples have been added, including 34 PPMI Cohort samples, 239 Steady PD Cohort samples, and the complete set of 259 samples for the new Sure PD Cohort. This brings the total to 8 cohorts with a combined total of 10,496 participants with a representative WGS sample and a Joint Genotyping Run that now includes 10,418 samples.
Proteomics Preview Data
This v2.5 release features a preview of the Targeted Proteomics dataset that will be combined with the Untargeted Proteomics dataset for the same participants from PDBP and PPMI cohorts in the v3.0 release. This Targeted Proteomics dataset preview contains eight unfiltered NPX files from four separate assays for matched Plasma and CSF samples. The four targeted proteomics assays are Cardiometabolic, Inflammation, Neurology, and Oncology. All samples (n=743) are from participants (n=212) with previously released clinical data, with matching WGS samples (n=205), with matching RNA samples (n=211), and with matching RNASeq samples from identical timepoints (n=484).
Composition by Cohort
BioFIND Data
Participants from the BioFIND cohort are represented in AMP PD clinical, RNA, and WGS data. Of 213 participants whose clinical records met AMP PD minimum clinical data criteria, 172 have corresponding WGS sample data (3 are represented by a linked WGS duplicate sample), 172 in the AMP PD joint genotyping dataset, 208 have corresponding RNA sample data, and 167 participants have corresponding samples in all three release data categories.
HBS Data
Participants from the Harvard Biomarkers Study (HBS) are represented in AMP PD clinical and WGS data. Of 1189 HBS participants whose clinical records met AMP PD minimum clinical data criteria, 1180 have corresponding WGS sample data (9 are represented by a linked WGS duplicate sample) and 1173 are represented in the AMP PD joint genotyping dataset.
PDBP Data
Participants from the Parkinson’s Disease Biomarkers Program (PDBP) are represented in AMP PD clinical, RNA, and WGS data. Of 1606 participants whose clinical records met AMP PD minimum clinical data criteria, 1505 have corresponding WGS sample data (7 are represented by a linked WGS sample), 1500 in the AMP PD joint genotyping dataset, 1484 have corresponding RNA sample data, and 1380 participants have corresponding samples in all three release data categories.
PPMI Data
Release 2.5 adds 34 WGS participant samples from the Parkinson’s Progression Markers Initiative (PPMI) bringing the total participants with representative data in AMP PD clinical, RNA, and WGS data to 1945. Of 1945 participants whose clinical records met AMP PD minimum clinical data criteria, 1809 have corresponding WGS sample data (7 are represented by a linked WGS duplicate sample), 1807 in the AMP PD joint genotyping dataset, 1582 have corresponding RNA sample data, and 1448 participants have corresponding samples in all three release data categories.
LBD Data
Participants from the Lewy Bodies Dementia (LBD) cohort are represented in AMP PD clinical and WGS data. Of 4586 HBS participants whose clinical records met AMP PD minimum clinical data criteria, 4579 have corresponding WGS sample data (7 are represented by a linked WGS duplicate sample) and 4579 are represented in the AMP PD joint genotyping dataset.
LCC Data
Participants from the LRRK2 Cohort Consortium (LCC) cohort are represented in AMP PD clinical and WGS data. Of 638 LCC participants whose clinical records met AMP PD minimum clinical data criteria, 599 have corresponding WGS sample data (39 are represented by a linked WGS duplicate sample) and 599 are represented in the AMP PD joint genotyping dataset.
Steady-PD
Release 2.5 adds 242 participants from the STEADY-PD3 cohort to complete the set. Of 334 total STEADY-PD3 cohort participants whose clinical records met AMP PD minimum clinical data criteria, 329 have corresponding WGS sample data (5 are represented by a linked WGS sample) and 334 are represented in the AMP PD joint genotyping dataset..
SURE-PD
In this Release 2.5, participants from the SURE-PD3 cohort are represented in AMP PD clinical and WGS data. Of 261 SURE-PD3 cohort participants whose clinical records met AMP PD minimum clinical data criteria, 259 have corresponding WGS sample data (2 are represented by a linked WGS sample) and 261 are represented in the AMP PD joint genotyping dataset.
Google Cloud Storage
Participant Data Products
- Table of all participants in all release data (n=10772)
- Table of all participants with minimum diagnosis information
- Table of all participant whole genome sequence samples in release data (n=10432)
- Table of all participant (n=3274) transcriptomics samples (n=8461) in release data
- Table of all participants who were included in more than one study and, therefore, appear in multiple clinical and genomics samples (n=79). For these genetically identical samples, a single WGS sample was selected with preference for the sample with higher mean coverage.
- Harmonized clinical data
- harmonized clinical data in 30 clinical forms as csv
- harmonized clinical per-form dictionary files as csv
WGS Data Products
- Table of all participant samples (n=10432) and processed file locations
- Single sample processed data: CRAM, gVCF, and GATK processing metrics (n=10432)
- Joint genotyping processed data: annotated variant vcf data (n=10418)
- Plink files: aggregated plink bfiles from all processed vcf data (n=10418)
- TOPMed joint genotyping processed data: annotated variant bcf data (n=4047)
RNA Data Products
- Table of all RNA participant (n=3274) samples (n=8461) and processed file locations
- Processed RNA sample data
- picard metrics: Aggregated per-sample alignment summary metrics, insert size metrics, and rna seq metrics. (n=8461)
- salmon quantification: Aggregated per-sample quantification estimates of the expression of transcripts and genes. Also available in matrix form. (n=8461)
- star align-reads: Aggregated per-sample Log.final.out outputs. (n=8461)
- feature counts: Aggregated per-sample featureCounts.tsv outputs. Also available in matrix form. (n=8461)
- plink genomes: Pairwise comparison of participants' RNA and WGS samples to detect sample contaminations, swaps and relatedness. (n=8461)
- multiqc reports: An html file containing visualizations from multiqc. Other multiqc artifacts are also available. (n=8461)
- sequencing metrics: Metrics from the sequencing provider. (n=8461)
- Processed RNA pooled sample data
- picard metrics: Aggregated per-sample alignment summary metrics, insert size metrics, and rna seq metrics. (n=192)
- salmon quantification: Aggregated per-sample quantification estimates of the expression of transcripts and genes. Also available in matrix form. (n=192)
- star align-reads: Aggregated per-sample Log.final.out outputs. (n=192)
- feature counts: Aggregated per-sample featureCounts.tsv outputs. Also available in matrix form. (n=n=192)
- multiqc reports: An html file containing visualizations from multiqc. Other multiqc artifacts are also available. (n=8461)
Google BigQuery
BigQuery Datasets
Participant Clinical Access BigQuery Dataset:
AMP PD Metadata Tables
amp_pd_participants
amp_pd_case_control
wgs_sample_inventory
amp_pd_participant_wgs_duplicates
wgs_gatk_joint_genotyping_samples
rna_sample_inventory
Clinical Participant Tables
Demographics, PD_Medical_History, Enrollment, Caffeine_history, Family_History_PD, Smoking_and_alcohol_history, LBD_Cohort_Clinical_Data, LBD_Cohort_Path_Data
Clinical Assessments Tables
Epworth_Sleepiness_Scale, MDS_UPDRS_Part_I,MDS_UPDRS_Part_II, MDS_UPDRS_Part_III, MDS_UPDRS_Part_IV, MMSE, MOCA, Modified_Schwab___England_ADL, PDQ_39, REM_Sleep_Behavior_Disorder_Questionnaire_Mayo, REM_Sleep_Behavior_Disorder_Questionnaire_Stiasny_Kolster, UPDRS, UPSIT
Clinical Bio Tables
Biospecimen_analyses_CSF_abeta_tau_ptau,Biospecimen_analyses_CSF_beta_glucocerebrosidase, Biospecimen_analyses_other, Biospecimen_analyses_SomaLogic_plasma, DaTSCAN_SBR, DaTSCAN_visual_interpretation, MRI, DTI
Participant Tier 2 BigQuery Dataset:
AMP PD Metadata Tables
amp_pd_participant_mutations
Clinical Participant Tables
Clinically_Reported_Genetic_Status
WGS BigQuery Dataset:
WGS Joint Genotyping Tables (n=10418)
gatk_passing_variants (per chromosome vcf tables)
gatk_variant_calling_detail_metrics
WGS Single Sample Variant Metrics Tables (n=10432)
gatk_variant_calling_summary_metrics
WGS Single Sample Alignment Metrics Tables (n=10432)
raw_wgs_metrics, wgs_metrics, preBqsr_selfSM
WGS Sample Metadata Tables (n=10432)
wgs_samples
wgs_sample_flags
RNA BigQuery Dataset:
RNA Sample Metadata Tables
rna_seq_samples
Picard Tables
alignment_summary_metrics, insert_size_metrics, rna_seq_metrics
Salmon Tables
quantification_genes, quantification_transcripts
Star Tables
star_metrics
FeatureCounts Tables
feature_counts
Plink Tables
genome_check_HW_MAF
Sequencing Tables
rna_quality_metrics
RNA Pools BigQuery Dataset:
RNA Pools Sample Metadata Tables
rna_seq_samples
Picard Tables
alignment_summary_metrics, insert_size_metrics, rna_seq_metrics
Salmon Tables
quantification_genes, quantification_transcripts
Star Tables
star_metrics
FeatureCounts Tables
feature_counts
V2 Release vs V2-5 Release Summary
Additions
- New cohort: Sure PD clinical and WGS samples
- Updated cohort: Steady PD clinical and WGS samples
- Updated cohort: PPMI clinical and WGS samples
- Added RNA Seq Pooled Samples
- New data type: Targeted Proteomics Preview
- Updated WGS: New per-chromosome Joint VCF
Changes
- Modified mutations table: Updated mutations data for v2 and v2-5 samples to replace null or "NA" values with valid data
- Removed WGS sample: SY-PDPR154JC1 is removed from WGS samples and Joint VCF, as valid data with incorrect cohort identification, to be reinserted in a subsequent release