News & Updates
AMP PD Release Notes - May 2022
With AMP PD’s latest update to the release 2.5 dataset, we have added new participant mutation data, which includes the APOE genotype, as well as updates to the targeted proteomics data, which was included for preview in release 2.5, to add additional metadata and qc information, and a correction to the MMSE form in the clinical data to correct a data issue affecting a small number of records.
Data Summary
Data Composition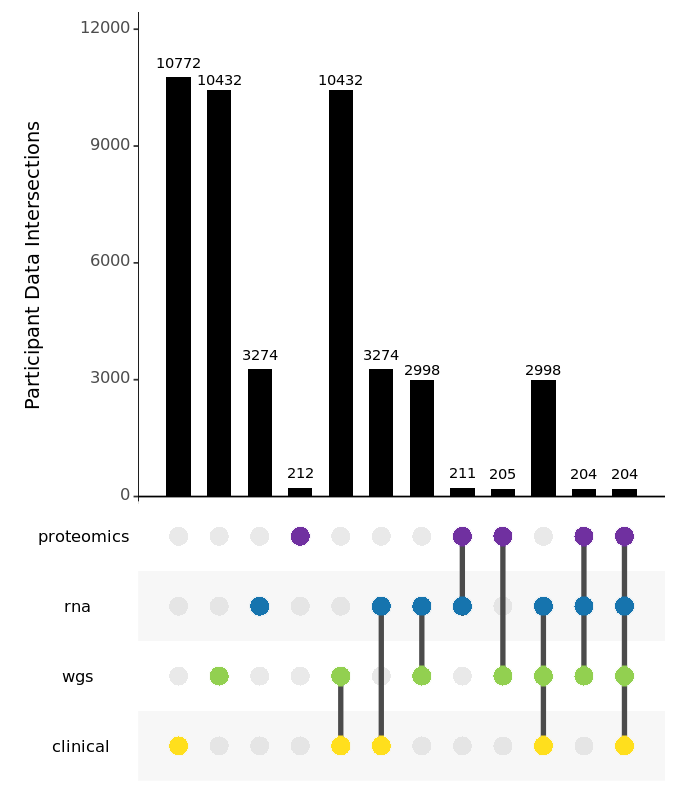
AMP PD's release 2.5 public dataset includes 10,772 participants from eight cohorts, including BioFIND, HBS, LBD, LCC, PDBP, PPMI, Steady PD, and Sure PD. AMP PD includes clinical data for all subjects, as well as whole genome sequencing data for 10,432 participants, transcriptomics data for 3,274 participants, and targeted proteomics data for 212 participants.
This release includes 2998 subjects with fully integrated clinical records, WGS samples, and RNA samples and, for 204 of these subjects, includes targeted proteomics data as well.
Data Availability
WGS data is available for download in Google Cloud Storage, including CRAM, gVCF, and GATK processing metrics for individual samples and annotated variant data in VCF and Plink format generated via joint genotyping. Transcriptomics data is also available for download, including picard metrics, salmon quantification, star align-reads, plink genomes, multiqc reports and sequencing metrics. Targeted proteomics data is available for download including normalized protein expression data.
Whole Genome Sequencing data, Transcriptomics data, Proteomics data and Clinical data is also available via SQL Query in Google BigQuery datasets.
Targeted Proteomics Data
For this release, AMP PD features an additional format for the preview release Targeted Proteomics dataset. This Targeted Proteomics dataset contains eight unfiltered NPX files from four separate assays for matched Plasma and CSF samples. The four targeted proteomics assays are Cardiometabolic, Inflammation, Neurology, and Oncology. All samples (n=743) are from participants (n=212) with previously released clinical data, with matching WGS samples (n=205), with matching RNA samples (n=211), and with matching RNASeq samples from identical timepoints (n=484).
Additional QC columns have been added to the data in order to provide additional information for flagged or passed samples. The released data has accompanying Terra notebooks, ranging from “Getting Started” notebooks which help users get familiar with the data and eventually assign case and control to samples, to QC notebooks to show users QC criteria used in flagging samples and analysis notebooks to perform simple data visualizations.
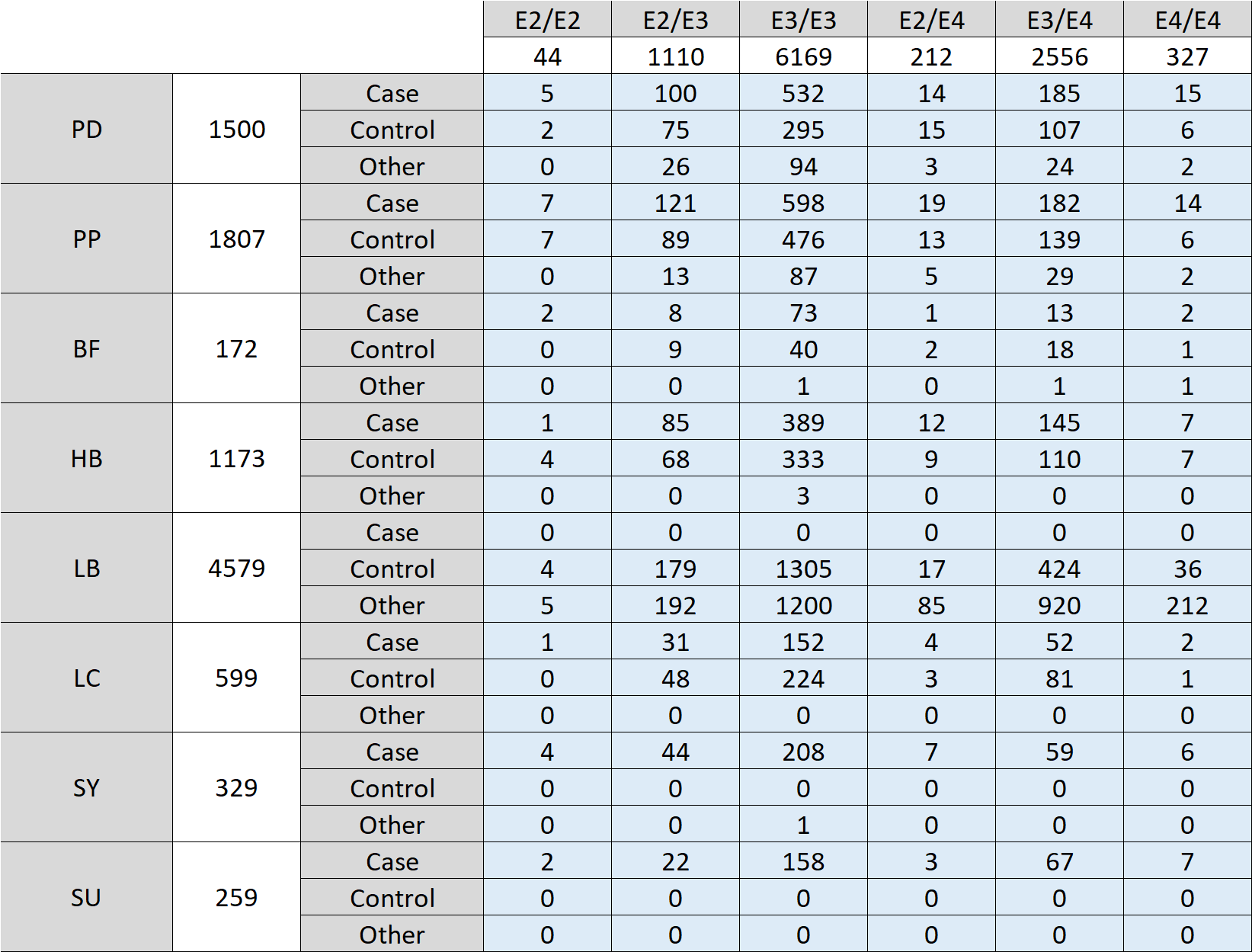
APOE Variant Data
Additional participant variant data is now available in AMP PD Tier 2 data identifying the APOE genotype for all AMP PD participants. Apolipoprotein (Apo) E is produced under the direction of the APOE gene and is one of five main types of blood lipoproteins (A-E). AMP PD has evaluated participant’s WGS data to determine what combination of APOE forms (genotype) is present. The APOE gene exists in three different forms (alleles) – e2, e3, and e4 – with e3 being the most common allele, found in 60% of the general population.
AMP PD’s public dataset includes 3,095 participants with at least one copy of the APOE E4 gene, and 327 participants with two copies.
MMSE Clinical Assessment Data Correction
A data error was identified in AMP PD’s previously released MMSE clinical assessment data. The values of mms108b_fold_in_half_score and mms109_close_eyes_score were transposed for 129 subjects in the HBS cohort. Updates have been made to correct this issue and the new MMSE data is available in the Release 2.5 clinical data in a file named ‘MMSE_corrected_20220514.csv’. For historical purposes, the original MMSE clinical data is available in a file named ‘MMSE_retracted.csv’. Similarly, in AMP PD’s Google BigQuery data, the MMSE data table has been renamed to MMSE_corrected_20220514 and the original data is available in the table named MMSE_retracted.
V2.5 Release vs Update Summary
Additions
New data type: Targeted Proteomics Preview Update
New Terra workspaces:
- Getting Started - Proteomics
- Proteomics QC & Analysis
New Tier 2 analysis data: Added APOE mutation genotype data
Changes
Modified clinical data: Corrected MMSE data